Note
Click here to download the full example code
1D bimodal example#
This example shows how to use lightkde.kde_1d and how it compares to
scipy.stats.gaussian_kde for a bimodal univariate case.
Import packages:
import matplotlib.pyplot as plt
import numpy as np
from scipy.stats import gaussian_kde, norm
from lightkde import kde_1d
Generate synthetic data from two univariate normal distributions:
np.random.seed(42)
sample = np.hstack((norm.rvs(size=2_000), 0.3 * norm.rvs(size=1_000) + 5))
Estimate kernel density using lightkde:
density_vec, x_vec = kde_1d(sample_vec=sample)
Estimate kernel density using scipy:
gkde = gaussian_kde(dataset=sample)
scipy_density_vec = gkde.evaluate(x_vec)
Plot the data against the kernel density estimates:
plt.plot(x_vec, density_vec, "--r", label="lightkde")
plt.plot(x_vec, scipy_density_vec, label="scipy.stats.gaussian_kde")
plt.hist(sample, bins=100, density=True, alpha=0.5, label="data")
plt.legend()
plt.show()
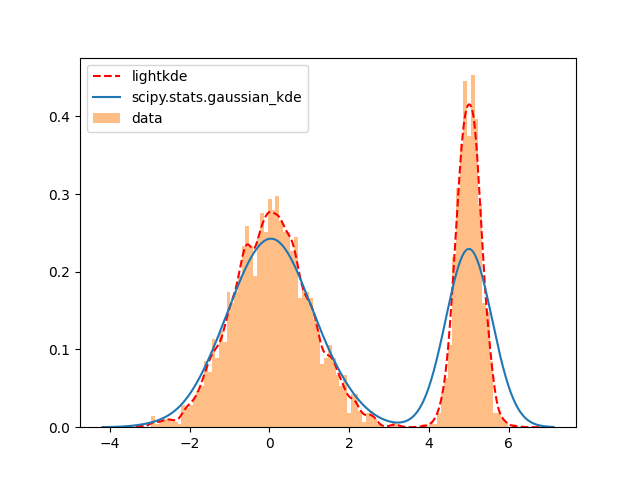
The scipy method oversmooths the kernel density and it is far
from the histogram of the data that it is expected to follow.
Total running time of the script: ( 0 minutes 1.833 seconds)